Transcriptional and post-transcriptional regulation of gene expression in the osteoblast
Transcriptional and post-transcriptional regulation of gene expression in the osteoblast
a. Regulation of bone formation by microRNA (V. Geoffroy)
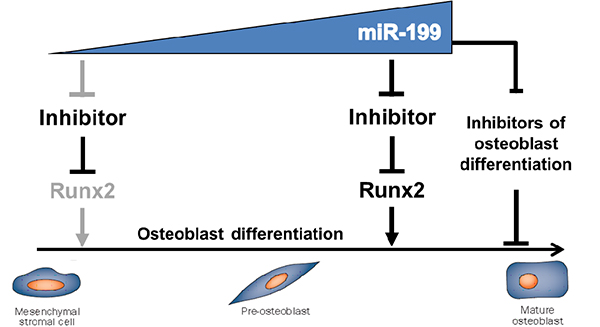
The regulation of gene expression is the result of a combination of transcriptional and post-transcriptional events. We have recently identified a new alternative for the regulation of osteoblast differentiation that involves microRNA and we found that Runx2, the key factor of bone development, is central in this new regulatory mechanism. We have shown that miR-199b-5p and miR468 cooperate to regulate the early stages of osteoblast differentiation (Cabon et al, 2012). These results together with previously published data indicate that miR-199-5p is not only capable of regulating osteoblast differentiation in vitro but could also control bone mass accrual in vivo (Watanabe et al, 2008). The main purpose of this project is to further characterize the role of miR-199a/b family in vitro and in vivo, in physiological and in bone loss situations. Transgenic mice will be generated in which the miR-199b-5p will be expressed in osteoblasts in order to evaluate its potential anabolic effect on bone. The expression and regulation of miR-199b-5p will be evaluated in bone during aging, after ovariectomy, in osteoblasts as well as in other bone cells (osteocytes and osteoclasts). Its effect on osteoblast proliferation, differentiation and apoptosis will be explored. We expect miR-199a/b family to have a positive role on bone formation in vivo.
For this purpose, identification of the upstream regulators of miR-199a/b family genes and their downstream targets will be of great interest since in vivo modulation of miR-199a/b may have a potential therapeutic application. The identification and validation of miR-199b-5p target genes will be performed using gain and loss of function using in vitro approaches. We have some evidence of a modification of the activity of the Wnt/βcatenin pathway after over-expression of miR-199b-5p. Several potential target genes have been identified in silico that include several effectors of the Wnt/βcatenin pathway such as GSK3β and NLK. We also aim to determine how transcriptional and post-transcriptional regulations cooperate to control Runx2 expression in vitro and in vivo. It is likely that a combination of transcription factors (TFs) and microRNAs are involved in the cell-specific regulation of Runx2 gene expression. For this study, reporter plasmids and reporter mouse lines will have to be generated in order to follow: 1) the upstream regulation of Runx2, and 2) the regulation of Runx2 by microRNA. This combinatory approach might help to explain how Runx2 is turned on and off in one given precursor cell or in osteoblasts. This project is part of the breakthrough therapeutic approaches, involving the application of microRNAs in the therapeutic field. In particular, miR-199b-5p could have potential therapeutic benefits in bone loss associated pathology such as osteoporosis. In the future, miR-199b-5p could also be used in the development of novel miRNA-based strategies for bone reconstruction of bone defects.
b. DNA methylation and gene expression (F. Jehan)
We intend to demonstrate that DNA methylation is an important regulator of gene expression in human bone. Methylation of DNA has been shown to control gene expression in physiological conditions (Kim et al., 2009), and is a regulator of important genes in bone metabolism including OPG and RANKL (Delgado-Calle et al., 2012a), SOST (Delgado-Calle et al., 2012b) and DKK1 (Kocemba et al., 2012).
We identified several methylated genes associated with changes in their expression in bone tissue: SOST and DKK1, key regulators of bone formation, and OPG and RANKL, genes that control bone remodeling. Using human osteoblastic cell lines and human bone samples, we will evaluate the impact of these changes in methylation on the dynamic structure of the promoter, i.e., changes in transcription factor binding and changes in the nucleosome structure and their impact on promoter activity.
Secondly, a combined analysis of DNA methylation and gene expression will be performed in order to identify other critical genes for which the differences in DNA methylation correlate with differences in gene expression. We will use the Infinium illumina array offering an ideal solution for genome-wide methylation profiling, to identify DNA methylation changes and to assess their relationship with changes in the expression of critical genes in human bone that will be quantified by microarray on the same samples.
Références bibliographiques
1. Adams GB,et al Nat Biotechnol. 2007 Feb;25(2):238-43. Epub 2007 Jan 21. Erratum in: Nat Biotechnol. 2007
2. Aug;25(8):944. Nat Biotechnol. 2008 Feb;26(2):241.
3. Chapurlat RD, Orcel P, 2008, Best Pract Res Clin Rheumatol. 22(1):55-69.
4. d’Alésio A et al. Hum Mol Genet. 200, 14:3539-48.
5. Delgado-Calle J et al, 2012, Epigenetics. 7:83-91.
6. Delgado-Calle J, et al, 2012, J Bone Miner Res. 27:926-37.
7. Fang Y,et al, 2005, Am. J. Hum. Genet. 77: 807-823.
8. Geoffroy V et al, 2002, Mol. Cell. Biol. 22 (17): 6222–6233.
9. Haÿ E et al, 2009 , Mol Cell Biol. 29, 4: 953-964.
10. Haÿ E et al, 2009, PloSOne 4(12):e8284.
11. Jehan F et al, J. Clin. Endoc. Metabol. 93: 4672–4682.
12. Kassem M, Marie PJ, 2011, Aging Cell, 1474-9726.
13. Kim MS et al, 2009, Nature, 461(7266):1007-12.
14. Kocemba KA et al, PLoS One. 2012;7(2):e30359. Epub 2012 Feb 17
15. Locker M et al, 2006, Cell Signal. 2006 May;18(5):628-39.
16. Marie PJ.2009. IBMS BoneKEy. 6(4):150-6.
17. Merciris Det al, 2007, Am J Pathol, 170(5):1676-1685
18. Ogita M et al, 2008, Endocrinology, 149(11):5713-23
19. Samee N et al, 2008, Am J pathol 173(3):773-80.
20. Samee N et al, 2009, J Cell Biochem, 107(5):865-72
21. Wu JY et al, 2009, J Bone Miner Res. 24(5):759-64.
22. Wu JY et al, 2011, J Clin Invest. 121(9):3492-504